ADNI Clinical Progression
ATRI Biostatistics
Progression.RmdADNI progression summaries using the ADNIMERGE R package
Description
This package vignette demonstrates how to use the ADNIMERGE package to generate progression summaries and plots with R and knitr.
Report generated from Progresssion.Rmd
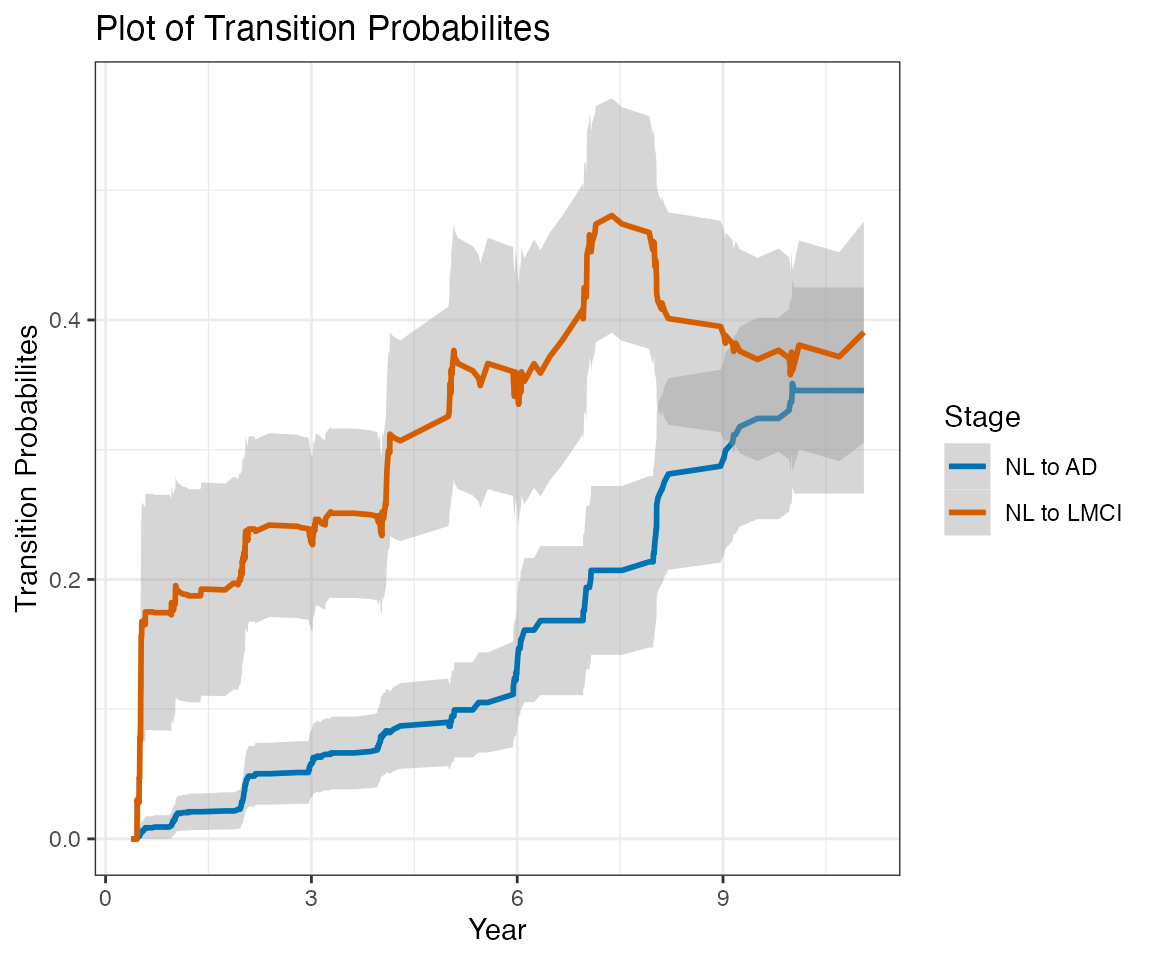
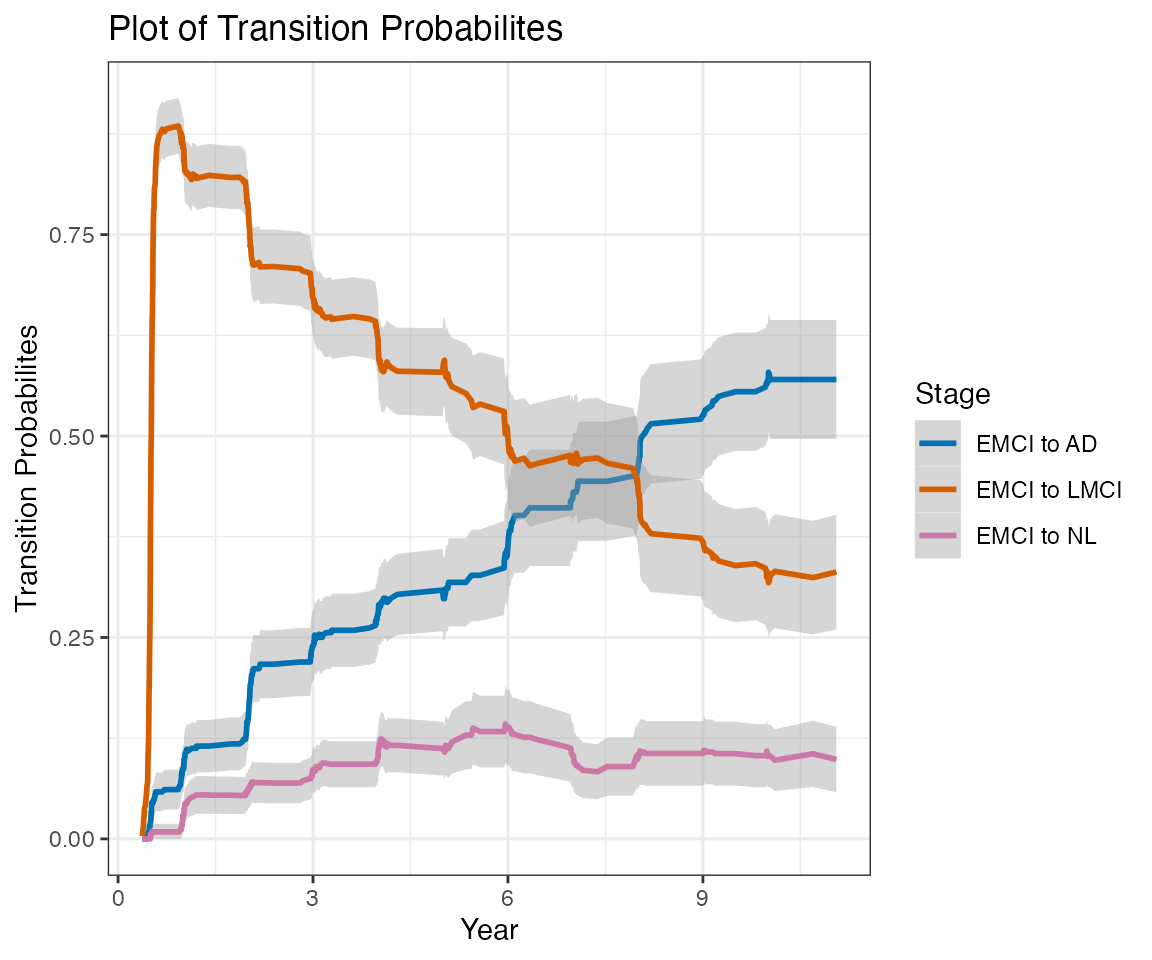
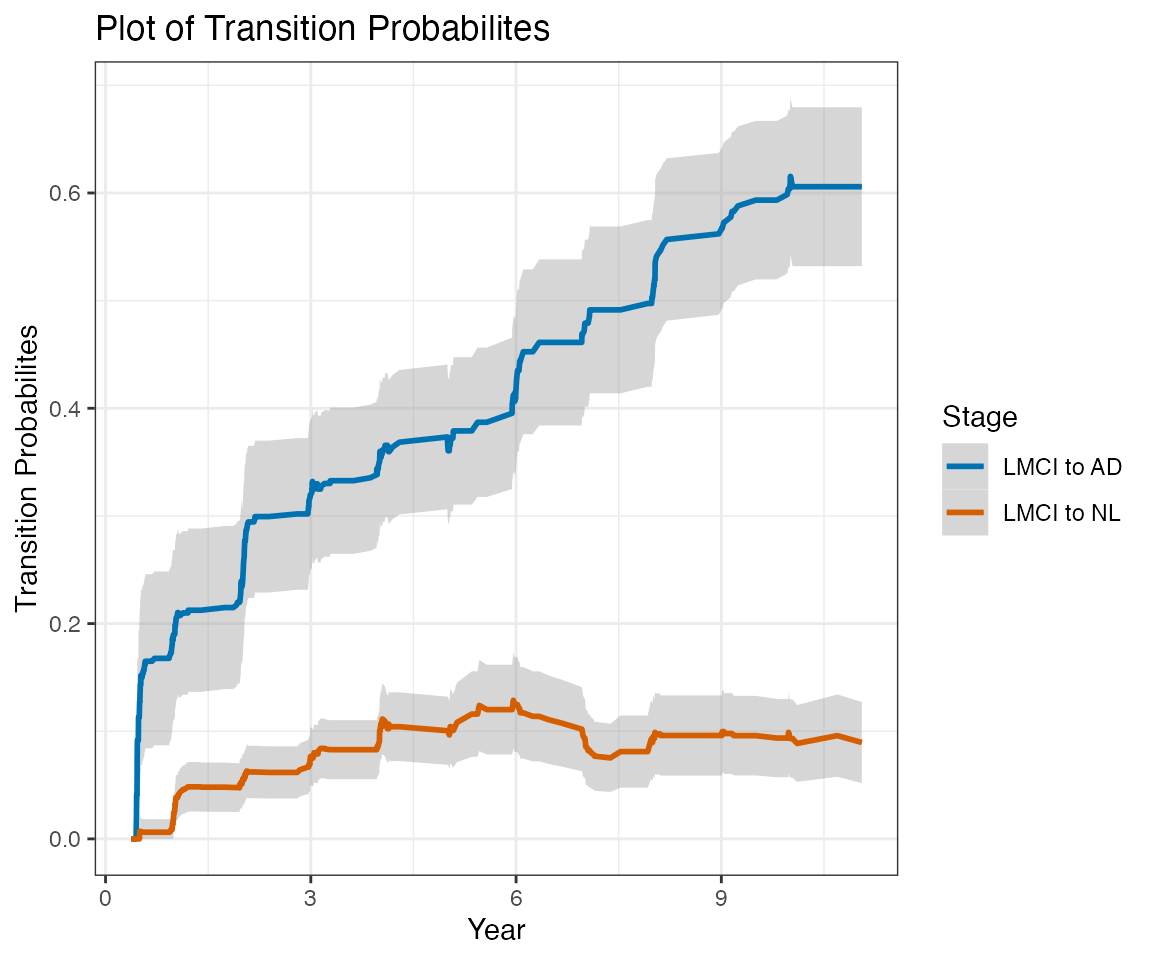
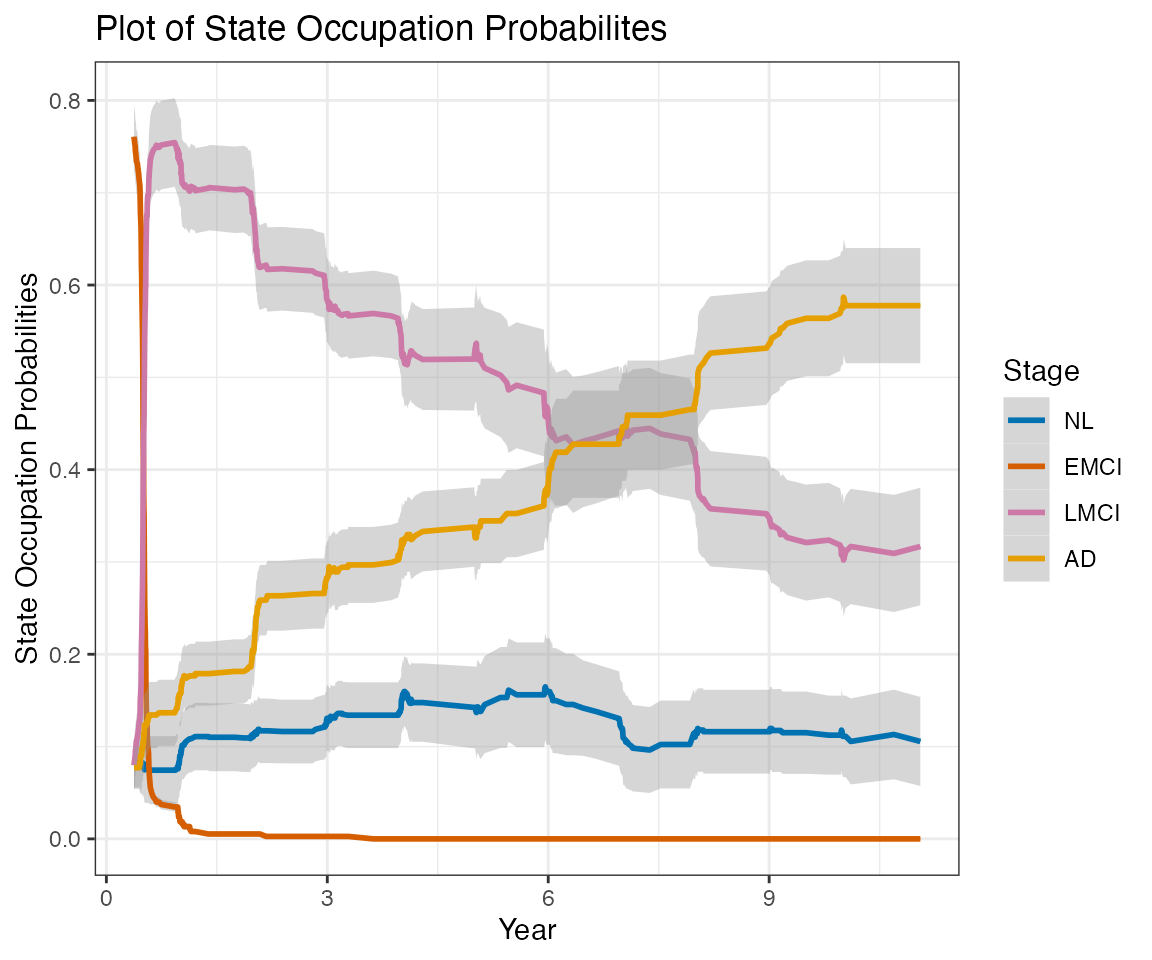
This report contains estimates of the ADNI conversion rates using a nonparametric multistate modeling approach (Nicole Ferguson, Somnath Datta, Guy Brock (2012). msSurv: An R Package for Nonparametric Estimation of Multistate Models. Journal of Statistical Software, 50(14), 1-24. http://www.jstatsoft.org/v50/i14/).
This report does not consider transition to EMCI at any followup visits. The label LMCI, when applied at followup visits, actually refer to any MCI (EMCI or LMCI).
Only subjects with at least one followup Diagnostic Summary are included in this report.
Load required R packages and functions
options(digits = 3)
library(knitr)
library(ADNIMERGE)
library(Hmisc)
library(msSurv)
library(zoo)
library(ggplot2)
theme_set(theme_bw())
source("http://adni.bitbucket.io/myfunctions.R")
cbbPalette <- c("#0072B2", "#D55E00", "#CC79A7", "#E69F00", "#56B4E9", "#009E73",
"#F0E442", "#000000")
scale_colour_discrete <- function(...) scale_colour_manual(..., values = cbbPalette)
scale_fill_discrete <- function(...) scale_fill_manual(..., values = cbbPalette)
No states eligible for entry distribution calculation.
Exit distributions calculated for states 2 .
Using bootstrap to calculate variances ... Transition counts (all intermediate transitions)
end.stage
start.stage NL EMCI LMCI AD Sum
NL 354 0 87 1 442
EMCI 2 0 281 4 287
LMCI 61 0 364 157 582
AD 0 0 19 282 301
Sum 417 0 751 444 1612Transition counts (first to last stage transitions)
end.stage
start.stage NL EMCI LMCI AD Sum
NL 348 0 48 14 410
EMCI 26 17 224 37 304
LMCI 11 0 155 93 259
AD 0 0 1 231 232
Sum 385 17 428 375 1205Transition probabilities (%)
Year 0 to 1
NL EMCI LMCI AD Sum
NL 79.048 0.00 18.99 1.96 100
EMCI 4.908 1.74 82.38 10.97 100
LMCI 4.336 0.00 74.92 20.74 100
AD 0.142 0.00 5.23 94.63 100
Sum 88.435 1.74 181.52 128.30 400Year 0 to 2
NL EMCI LMCI AD Sum
NL 71.28 0.000 23.89 4.84 100
EMCI 6.98 0.697 71.20 21.13 100
LMCI 6.22 0.000 64.34 29.44 100
AD 0.37 0.000 8.03 91.60 100
Sum 84.84 0.697 167.46 147.01 400